← Back to Projects

This project is a sub-project of AgCloud

Ground
Mentored by: Vast Data
Ground - Cloud-based platform for agricultural data management and analytics

Apache Kafka
Apache Flink
Microservices
Docker
Prometheus
Grafana
Airflow
PostgreSQL
PostGIS
MinIO
FastAPI
CI/CD
pytest
Python
PyQt6
PostgreSQL
MQTT
Description
Ground sub-project of AgCloud. A comprehensive cloud platform for managing agricultural operations, data, and analytics. Provides centralized storage, processing, and visualization of farm data including crop monitoring, weather integration, equipment management, and predictive analytics. Features include multi-tenant architecture, real-time dashboards, and API integrations.
Mentors
R
Renana Grilus
VAST Data
O
Orian Ohayon
Software engineer
VAST Data
Team Members
Cohort: Data Science Bootcamp 2025 (Data)
Responsibilities:
Schemas & Validation: Built JSON/Protobuf schemas for sensor readings with validation scripts, folder-level checks, and CI integration (GitHub Actions + Spark cleaning job).
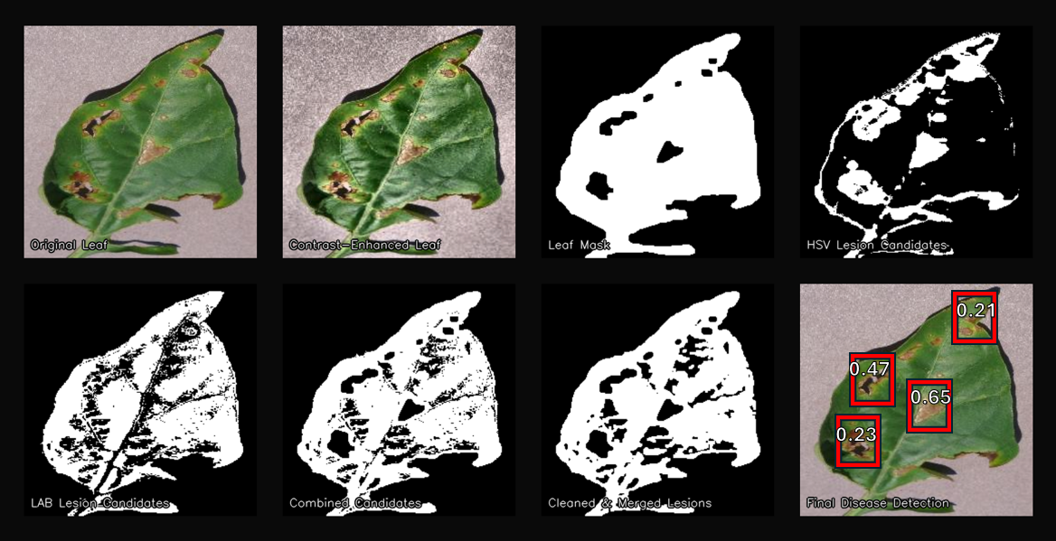
Graph-Based Background Removal: Implemented classical segmentation for foreground masks and cut-outs, with CLI/API wrapper and post-processing.
Disease Anomaly & Worsening Detection: Developed offline pipeline for detecting anomalies and worsening trends in disease data using statistical baselines, Z-score/IQR/CUSUM, alert deduplication, and Postgres logging.
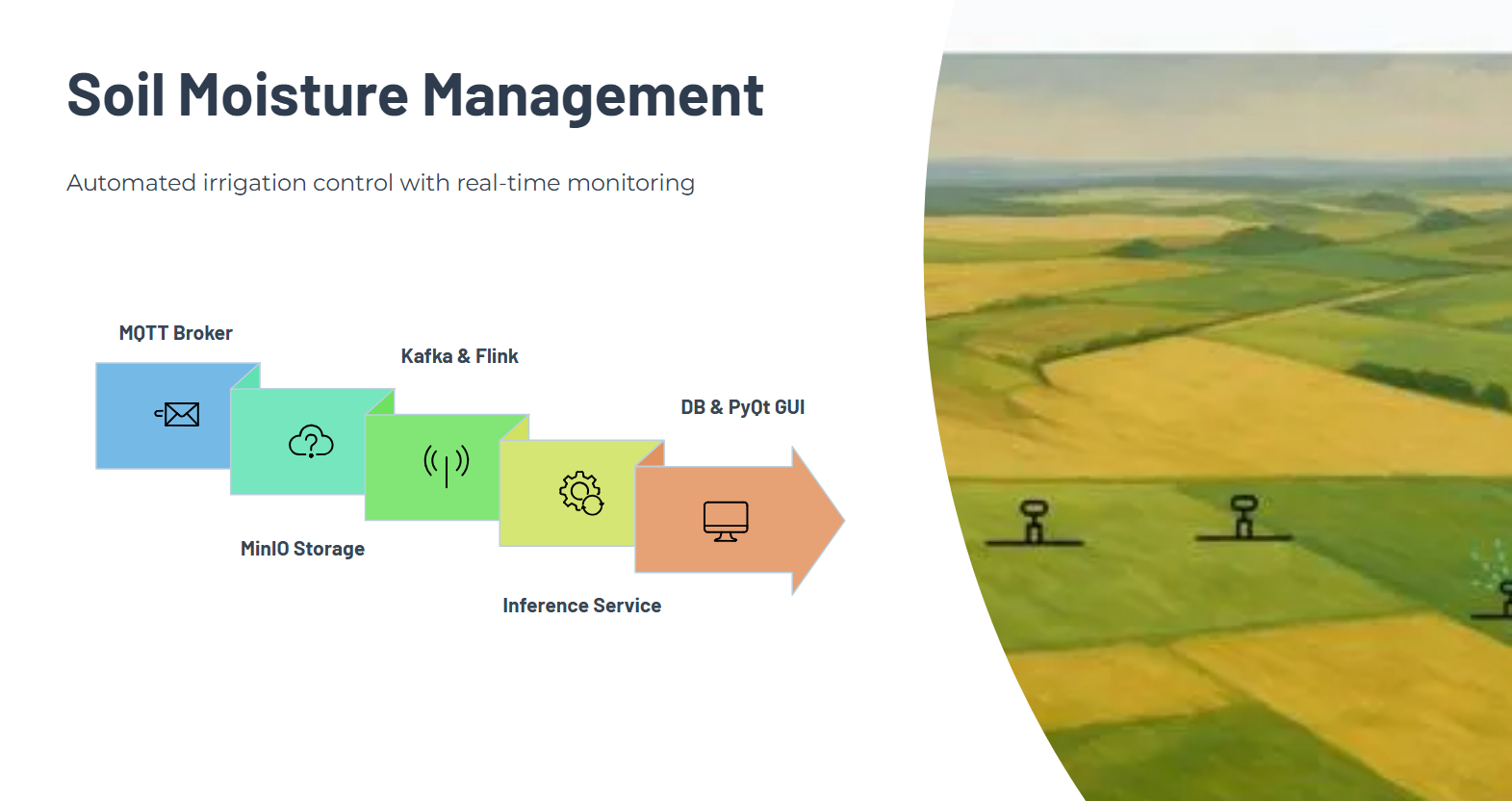
Soil Moisture Detection Pipeline: Built real-time CV pipeline (FastAPI + MobileNetV3) for wet/dry classification with threshold-based alerts, Kafka messaging, DLQ retries, and Postgres event logging.
End-to-End Integration: Connected full pipeline from MinIO image ingestion → Kafka → Flink processing → inference service → DB updates → PyQt GUI visualization of zones, history, and irrigation status.
PyQt GUI: Created interactive map showing all sprinklers, active zones, last images, history, and parameter updates
...and more contributions not listed here
Responsibilities:
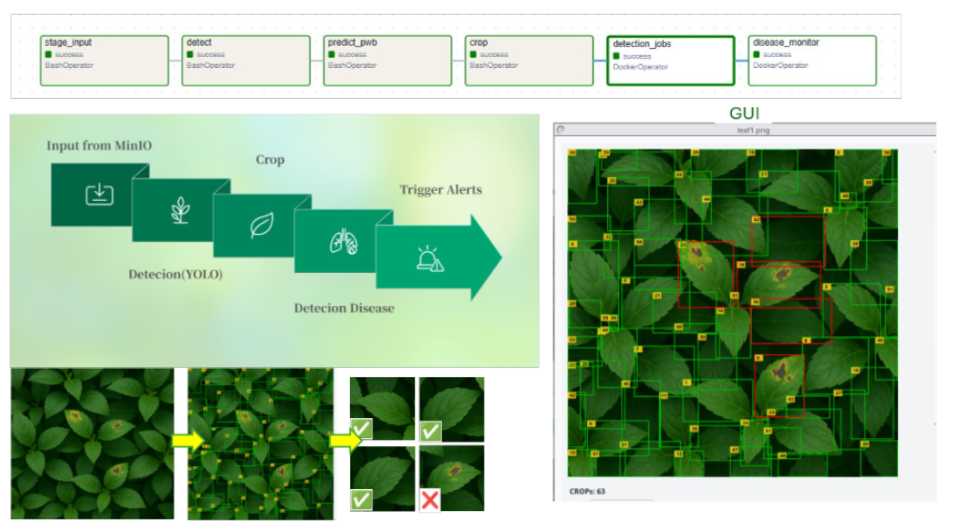
End-to-End Leaf Disease Detection Pipeline A comprehensive pipeline that ingests raw leaf imagery, performs automated preprocessing, applies YOLO-based leaf detection, extracts individual patches, classifies disease symptoms, generates structured metadata, and stores all outputs for real-time analytics and downstream processing.
Kafka Stream + Embedding Service Workflow A streaming workflow in which a Kafka consumer ingests imagery-notification events, validates and normalizes them, batches requests to a CLIP-based gRPC embedding service for vector generation, enriches each message with embeddings, and publishes the enhanced events to downstream Kafka topics with DLQ handling.
Responsibilities:
End-to-End Leaf Disease Service (MinIO → OpenCV/Model → PostgreSQL): Built an end-to-end Python service that pulls leaf images from MinIO, runs an OpenCV disease detector, and writes structured results into PostgreSQL, turning raw images into traceable leaf-disease reports in the AgCloud pipeline.
Leaf Disease Detection Logic (Multi-stage ML Training): Built a three-stage ResNet18 training pipeline (PlantVillage → PlantDoc fine-tuning) using PyTorch, Albumentations and MixUp, to classify each leaf as healthy or sick and assign a specific disease class with robust performance on real-field images.
Leaf Disease Dashboard (Desktop + Grafana Integration): Developed a PyQt6 “LeafDiseaseView” dashboard that queries leaf reports from PostgreSQL via a REST API, computes key KPIs, ranks devices and diseases, and embeds a Grafana drill-down view per disease and date range for interactive field-level analytics.
Kafka + Flink Automated Test Lab with PyTest & Testcontainers: Implemented an automated Kafka+Flink “test lab” using PyTest and Testcontainers that spins up ephemeral clusters, streams hundreds of test images through the pipeline, and verifies exactly-once processing with high branch coverage as part of the CI pipeline.